RDFpro usage example (SemDev paper)
We describe here an example of using RDFpro for integrating RDF data from Freebase, GeoNames and DBpedia (version 3.9) in the four languaged EN, ES, IT and NL, performing smushing, inference, deduplication and statistics extraction.
This example is taken from the ISWC SemDev 2014 paper and here we provide further details included the concrete RDFpro commands necessary to carry out the integration task. Note, however, that the numbers here reported (collected after repeating the processing in January 2015) are different from the ones in the paper, as both the accessed data sources and RDFpro have changed in the meanwhile (more data available, faster RDFpro implementation).
Data download and selection
We assume the goal is to gather and integrate data about entities (e.g., persons, location, organizations) from the three Freebase, GeoNames and DBpedia sources. In a first selection step we choose the following dump files as input for our integration process:
-
the Freebase complete RDF dataset, containing over 2.6 billions triples.
-
the GeoNames data file, ontology and schema mappings w.r.t. DBpedia
-
the following DBpedia 3.9 files for the four EN, ES, IT, NL DBpedia chapters:
- DBpedia ontology
- article categories, EN, ES, IT, NL
- category labels, EN, ES, IT, NL
- external links, EN, ES, IT, NL
- geographic coordinates, EN, ES, IT, NL
- homepages, EN, ES, IT
- image links, EN, ES, IT
- instance types, EN, EN (heuristics), ES, IT, NL
- labels, EN, ES, IT, NL
- mapping-based properties, EN (cleaned), ES, IT, NL
- short abstracts, EN, ES, IT, NL
- SKOS categories, EN, ES, IT, NL
- Wikipedia links, EN, ES, IT, NL
- interlanguage links, EN, ES, IT, NL
- DBpedia IRI - URI owl:sameAs links, EN, ES, IT, NL
- DBpedia - Freebase owl:sameAs links, EN, ES, IT, NL
- DBpedia - GeoNames owl:sameAs links, EN, ES, IT, NL
- person data, EN
- PND codes, EN
-
vocabulary definition files for FOAF, SKOS, DCTERMS, WGS84, GEORSS
Basically, we import all Freebase and GeoNames data as they only provide global dump files, while we import selected DBpedia dump files leaving out files for Wikipedia inter-page, redirect and disambiguation links and page and revision IDs/URIs (as not relevant in this scenario), raw infobox properties (as lower-quality if compared to mapping-based properties) and extended abstract (as too long).
Bash script download.sh can be used to automatize the download of all the selected dump files, placing them in multiple directories vocab (for vocabularies), freebase, geonames, dbp_en (DBpedia EN), dbp_es (DBpedia ES), dbp_it (DBpedia IT) and dbp_nl (DBpedia NL).
Data processing (single steps)
Processing with RDFpro involves the six steps shown in the figure below. These steps can be executed individually by invoking rdfpro six times, as described next. Bash script process_single.sh can be used to issue these commands.
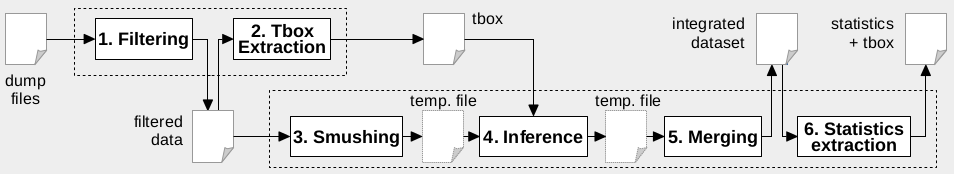
Step 1 Filtering
rdfpro { @read metadata.trig , \
@read vocab/* @transform '=c <graph:vocab>' , \
@read freebase/* @transform '-spo fb:common.topic fb:common.topic.article fb:common.topic.notable_for
fb:common.topic.notable_types fb:common.topic.topic_equivalent_webpage <http://rdf.freebase.com/key/*>
<http://rdf.freebase.com/ns/common.notable_for*> <http://rdf.freebase.com/ns/common.document*>
<http://rdf.freebase.com/ns/type.*> <http://rdf.freebase.com/ns/user.*> <http://rdf.freebase.com/ns/base.*>
<http://rdf.freebase.com/ns/freebase.*> <http://rdf.freebase.com/ns/dataworld.*>
<http://rdf.freebase.com/ns/pipeline.*> <http://rdf.freebase.com/ns/atom.*> <http://rdf.freebase.com/ns/community.*>
=c <graph:freebase>' , \
@read geonames/*.rdf .geonames:geonames/all-geonames-rdf.zip \
@transform '-p gn:childrenFeatures gn:locationMap
gn:nearbyFeatures gn:neighbouringFeatures gn:countryCode
gn:parentFeature gn:wikipediaArticle rdfs:isDefinedBy
rdf:type =c <graph:geonames>' , \
{ @read dbp_en/* @transform '=c <graph:dbp_en>' , \
@read dbp_es/* @transform '=c <graph:dbp_es>' , \
@read dbp_it/* @transform '=c <graph:dbp_it>' , \
@read dbp_nl/* @transform '=c <graph:dbp_nl>' } \
@transform '-o bibo:* -p dc:rights dc:language foaf:primaryTopic' } \
@transform '+o <*> _:* * *^^xsd:* *@en *@es *@it *@nl' \
@transform '-o "" ""@en ""@es ""@it ""@nl' \
@write filtered.tql.gz
Downloaded dump files are filtered to extract desired RDF quads and place them in separate graphs to track provenance. A metadata file is added to link each graph to the URI of the associated source (e.g. Freebase). The command above shows how a parallel and sequence composition of @read and @filter can be used to process in a single step a number of RDF files, applying filtering both to separate file groups and globally. Some notes on the implemented filtering rules:
- Freebase filtering aims at removing redundant http://rdf.freebase.com/key/ triples and triples belonging to Freebase specific domains (e.g., users, user KBs, schemas, …). We also remove triples of limited informative value.
- GeoNames filtering aims at removing triples that are uninformative (rdfs:isDefinedBy, rdf:type), redundant (gn:countryCode, gn:parentFeature, gn:wikipediaArticle, the latter providing the same information of links to DBpedia) or that points to auto-generated resources for which there is no data in the dump (gn:childrenFeatures, gn:locationMap, gn:nearbyFeatures, gn:neighbouringFeatures).
- DBpedia filtering aims at removing triples that have limited informative value (dc:rights, dc:language for images and Wikipedia pages) or that are redundant (foaf:primaryTopic and all triples in the bibo namespace).
- the global filtering ("ol -'' o@ +'en' +'es' +'it' +'nl' -* o^ +xsd -*") aims at removing literals with a language different from en, es, it and nl, literals that are empty and literals with a datatype not in the XML Schema vocabulary.
- the -r "cu '<GRAPH_URI>'" options serve to place filtered triples in different graphs, so that we can keep track of which source they come from
Step 2 TBox extraction
rdfpro @read filtered.tql.gz \
@tbox \
@transform '-o owl:Thing schema:Thing foaf:Document bibo:* con:* -p dc:subject foaf:page dct:relation bibo:* con:*' \
@write tbox.tql.gz
TBox quads are extracted from filtered data and stored, filtering out unwanted top level classes (owl:Thing, schema:Thing, foaf:Document) and vocabulary alignments (to bibo and con terms and dc:subject).
Step 3 Smushing
rdfpro @read filtered.tql.gz \
@smush '<http://dbpedia>' '<http://it.dbpedia>' '<http://es.dbpedia>' \
'<http://nl.dbpedia>' '<http://rdf.freebase.com>' '<http://sws.geonames.org>' \
@write smushed.tql.gz
Filtered data is smushed so to use canonical URIs for each owl:sameAs equivalence class, producing an intermediate smushed file. Note the specification of a ranked list of namespaces for selecting the canonical URIs.
Step 4 Inference
rdfpro @read smushed.tql.gz \
@rdfs -c '<graph:vocab>' -e rdfs4a,rdfs4b,rdfs8 -d tbox.tql.gz \
@transform '-o owl:Thing schema:Thing foaf:Document bibo:* con:* -p dc:subject foaf:page dct:relation bibo:* con:*' \
@write inferred.tql.gz
The deductive closure of smushed data is computed and saved, using the extracted TBox and excluding RDFS rules rdfs4a, rdfs4b and rdfs8 (and keeping the remaining ones) to avoid inferring uninformative X rdf:type rdfs:Resource quads. The closed TBox is placed in graph <graph:vocab>. A further filtering is done to be sure that no unwanted triple is present in the result dataset due to inference.
Step 5 Merging
rdfpro @read inferred.tql.gz \
@unique -m \
@write dataset.tql.gz
Quads with the same subject, predicate and object are merged and placed in a graph linked to the original sources to track provenance (note the use of the -m option).
Step 6 Statistics extraction
rdfpro { @read tbox.tql.gz , @read dataset.tql.gz @stats } @write statistics.tql.gz
VOID statistics are extracted and merged with TBox data, forming an annotated ontology that documents the produced dataset.
Data processing (aggregate steps)
The 6 steps previously listed can be also aggregated to reduce overhead for writing and reading back intermediate files, exploiting RDFpro capability to arbitrarily compose processors and write intermediate results. In particular, steps 1-2 can be aggregated as follows:
rdfpro { @read metadata.trig , \
@read vocab/* @transform '=c <graph:vocab>' , \
@read freebase/* @transform '-spo fb:common.topic fb:common.topic.article fb:common.topic.notable_for
fb:common.topic.notable_types fb:common.topic.topic_equivalent_webpage <http://rdf.freebase.com/key/*>
<http://rdf.freebase.com/ns/common.notable_for*> <http://rdf.freebase.com/ns/common.document*>
<http://rdf.freebase.com/ns/type.*> <http://rdf.freebase.com/ns/user.*> <http://rdf.freebase.com/ns/base.*>
<http://rdf.freebase.com/ns/freebase.*> <http://rdf.freebase.com/ns/dataworld.*>
<http://rdf.freebase.com/ns/pipeline.*> <http://rdf.freebase.com/ns/atom.*> <http://rdf.freebase.com/ns/community.*>
=c <graph:freebase>' , \
@read geonames/*.rdf .geonames:geonames/all-geonames-rdf.zip \
@transform '-p gn:childrenFeatures gn:locationMap
gn:nearbyFeatures gn:neighbouringFeatures gn:countryCode
gn:parentFeature gn:wikipediaArticle rdfs:isDefinedBy
rdf:type =c <graph:geonames>' , \
{ @read dbp_en/* @transform '=c <graph:dbp_en>' , \
@read dbp_es/* @transform '=c <graph:dbp_es>' , \
@read dbp_it/* @transform '=c <graph:dbp_it>' , \
@read dbp_nl/* @transform '=c <graph:dbp_nl>' } \
@transform '-o bibo:* -p dc:rights dc:language foaf:primaryTopic' } \
@transform '+o <*> _:* * *^^xsd:* *@en *@es *@it *@nl' \
@transform '-o "" ""@en ""@es ""@it ""@nl' \
@write filtered.tql.gz \
@tbox \
@transform '-o owl:Thing schema:Thing foaf:Document bibo:* con:* -p dc:subject foaf:page dct:relation bibo:* con:*' \
@write tbox.tql.gz
Similarly, steps 3-6 can be aggregated in a single macro-step:
rdfpro @read filtered.tql.gz \
@smush '<http://dbpedia>' '<http://it.dbpedia>' '<http://es.dbpedia>' \
'<http://nl.dbpedia>' '<http://rdf.freebase.com>' '<http://sws.geonames.org>' \
@rdfs -c '<graph:vocab>' -e rdfs4a,rdfs4b,rdfs8 -d tbox.tql.gz \
@transform '-o owl:Thing schema:Thing foaf:Document bibo:* con:* -p dc:subject foaf:page dct:relation bibo:* con:*' \
@unique -m \
@write dataset.tql.gz \
@stats \
@read tbox.tql.gz \
@write statistics.tql.gz
Bash script process_aggregated.sh can be used to issue these two commands.
Results
The table below reports the results of executing the processing steps individually and aggregated on an Intel Core I7 860 machine with 16 GB RAM and a 500GB 7200RPM hard disk, using pigz and pbzip2 as compressors/decompressors and sort -S 4096M --batch-size=128 --compress-program=pigz as the sort command. Smushing and inference add duplicates that are removed with merging. TBox extraction and filtering are fast, while other steps are slower because complex or due to the need to sort data or process it in multiple passes. The aggregation of processing steps leads to a sensible reduction of the total processing time from 17500 s to 10905 s (note: times were respectively 18953 s and 13531 s in July 2014 test).
| Processing step | Input size | Output size | Throughput | Time [s] |
|||
|---|---|---|---|---|---|---|---|
| [Mquads] | [MiB] | [Mquads] | [MiB] | [Mquads/s] | [MiB/s] | ||
| Step 1 - Filtering | 3175 | 31670 | 770 | 9871 | 0.76 | 7.55 | 4194 |
| Step 2 - TBox extraction | 770 | 9871 | <1 | ~1 | 1.87 | 23.95 | 412 |
| Step 3 - Smushing | 770 | 9871 | 800 | 10538 | 0.34 | 4.36 | 2265 |
| Step 4 - Inference | 800 | 10539 | 1691 | 15884 | 0.21 | 2.79 | 3780 |
| Step 5 - Merging | 1691 | 15884 | 964 | 9071 | 0.40 | 3.73 | 4254 |
| Step 6 - Statistics extraction | 964 | 9072 | <1 | ~3 | 0.37 | 3.50 | 2595 |
| Steps 1-2 aggregated | 3175 | 31670 | 770 | 9872 | 0.74 | 7.34 | 4315 |
| Steps 3-6 aggregated | 770 | 9872 | 964 | 9080 | 0.12 | 1.50 | 6590 |


